Unique AI-based super-fast search in chemical space
Scientists:
Scientists: Miroslav Lžičař, Hamza Gamouh, Martin Culka, Jan Skácel, Jan Macek
Challenge
The ability to rapidly search and analyze large databases of molecules for similar compounds have revolutionized drug discovery and chemical informatics. Searching large databases using traditional means is, however, extremely slow, and therefore intractable. To address those shortcomings, we developed a new fast and scalable tool: CHEESE (Chemical Embeddings Search Engine) for searching very large chemical spaces.
The AI-based tool learns molecule representations and has the potential to be trainable and flexible to many molecule similarity aspects (2D, 3D, ...). Furthermore, the learned molecule representations can be leveraged for multiple downstream tasks such as molecular property prediction.
Technology
To construct our tool, we used the ZINC15 database (700 million molecules). We trained an AI model to learn three similarity metrics: Morgan 2D Fingerprint similarity, 3D Shape similarity and Electrostatic similarity. The 3D shape similarity metric between two molecules is based on the 3D surface overlap of the best aligned pair of randomly generated conformers. The Electrostatic similarity is based on the best overlap of the electrostatic potential (ESP) between aligned pairs of conformers. The above metrics should preserve the protein-ligand binding properties of query molecules. Searching the whole ZINC15 database using the exact shape and electrostatic similarity metrics is intractable. Our method presents a huge advantage since it enables to search the database using different similarity metrics in few seconds. Moreover, the learned representations can be visualized together and allow for intuitive neighborhood exploration of the query.
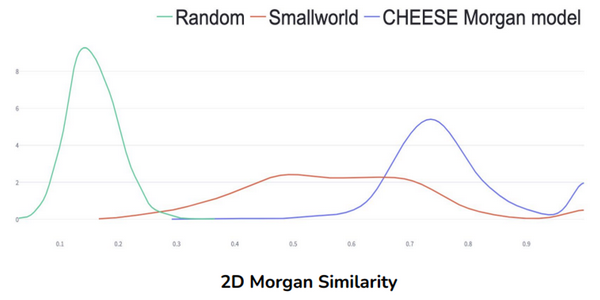
CHEESE Evaluation. Comparison of our models against SOTA molecular search on enumerative databases called Smallworld (used in ZINC22) and random molecule retrieval. 100 search queries with 30 results were performed using each method. Our models are clearly better than random search since they can retrieve much more similar molecules. They are also beating Smallworld on 2D Morgan and 3D Electrostatic search.
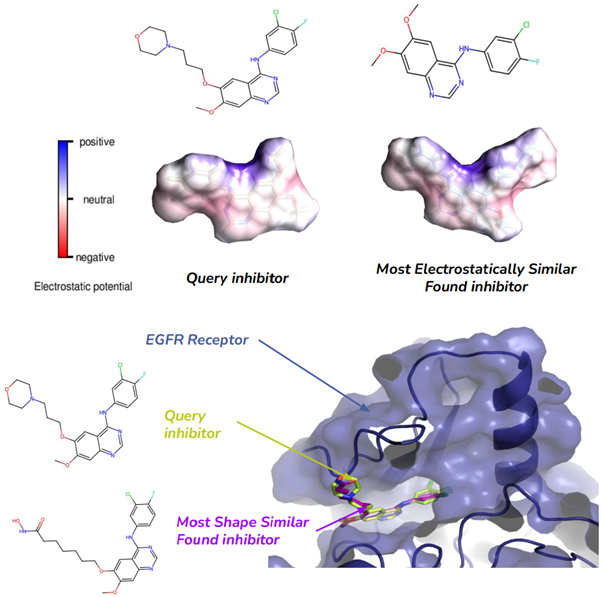
Case Study: Finding Inhibitors. To show the potential application of our tool, we performed a case study where we worked with the human Epidermal Growth Factor Receptor (EGFR). The EGFR has more than 3000 inhibitors in the ZINC15 database. We searched 100 randomly selected inhibitors using our tool, and we were able to find at least one new inhibitor for 14 of the query inhibitors. Interestingly, we have found 5 new inhibitors of one particular inhibitor query. Moreover, some newly found inhibitors were highly similar to the query in 3D Shape Similarity and Electrostatic Similarity. This shows that our tool is able to find molecules with similar binding properties and is very promising to find new inhibitors for a target.
Coming features include integration in a virtual screening pipeline with property prediction, molecule generation & docking; target representation to navigate to more promising chemical regions; extend to further relevant use cases of the molecular embeddings.
Commercial opportunity
The AI-driven drug discovery market was valued at $627.1 million in 2021 and is projected to grow to $4,197.5 million by 2028. This growth represents a compound annual growth rate (CAGR) of 41.5% from 2022 to 2028.
CHEESE, as well as other AI-driven methods for drug discovery, are being actively developed by us to become the world-leading technology for large chemical space exploration.
Development status
CHEESE search engine has been deployed at cheese.themama.ai and is ready to use. Other catalogs of compounds as well as novel features will be added subsequently. We further offer customization of the engine to customers’ databases and their needs.
Categories
Computational chemistry; CADD; Artificial intelligence
Keywords
artificial intelligence machine learning large chemical space similarity search virtual screening